Motor neuron atlas maps a path to understanding adult-onset diseases of the nervous system
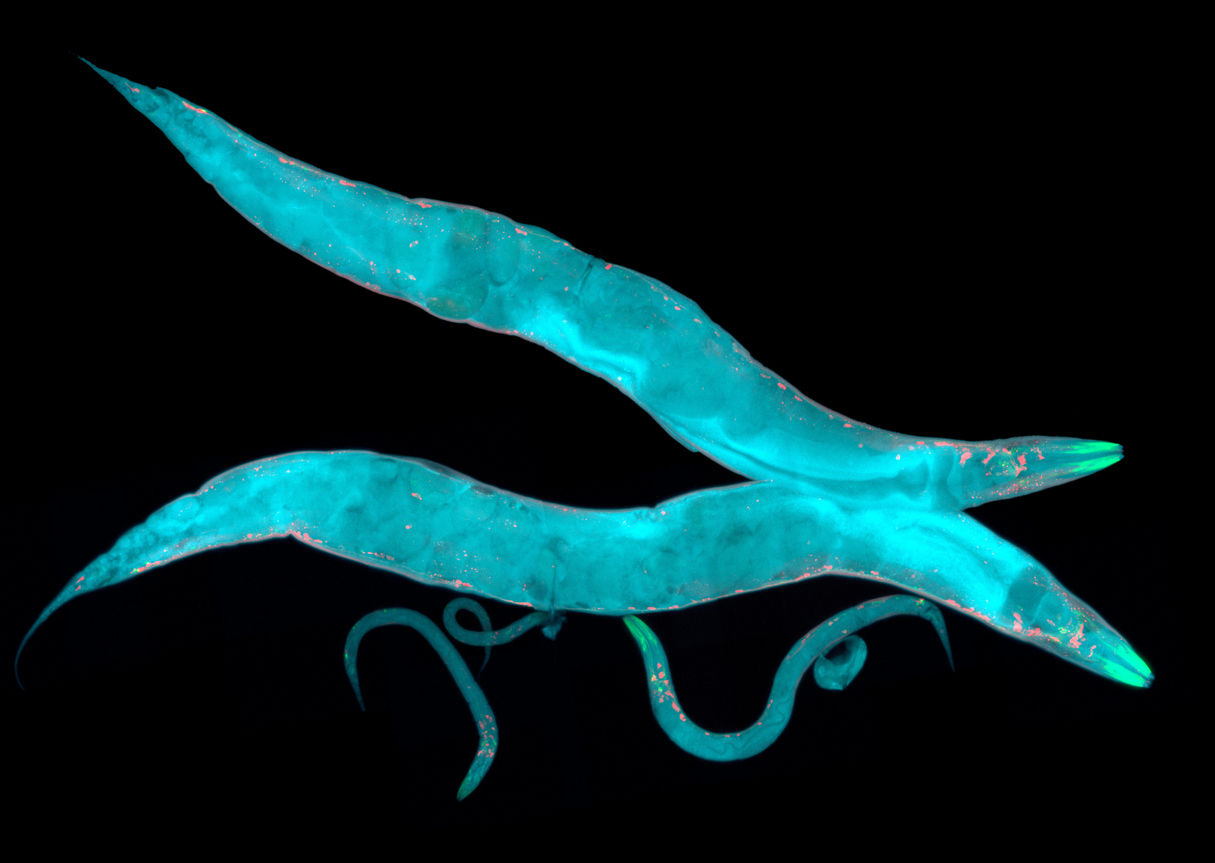
Single-cell RNA-sequencing by UChicago and Vanderbilt researchers builds a complete map of adult motor neurons in a classic animal model used to understand the fundamentals of the nervous system.
Motor neurons, which connect the central nervous system to muscles and control movement, are among the most extensively studied nerve cells in biology. Much of what scientists know about motor neurons happens during their developmental stages, including their birth, migration, and formation of synapses. Many of the most devastating diseases of the motor nervous system, however, such as amyotrophic lateral sclerosis (ALS), begin during adulthood when the motor neurons are fully formed.
A new study by neurobiologists from the University of Chicago and Vanderbilt University hopes to bridge this gap by creating a complete atlas of molecular and genetic activity of adult motor neurons in the C. elegans roundworm, a classic animal model used to understand the fundamentals of the nervous system. This new atlas, when combined with existing data on connectivity among C. elegans neurons, describes a surprising diversity of different motor neuron subtypes (or subclasses), and maps out organizing principles that are conserved from invertebrates to mammals.
“If we want to understand and treat adult-onset diseases of the nervous system, we must have a blueprint of how the system is built,” said senior author Paschalis Kratsios, PhD, Associate Professor of Neurobiology and co-Director of the Center for Motor Neuron Disease (CMND) at the University of Chicago. “This paper is actually the first step in understanding – at the molecular level – a clinically relevant cell type that is attacked by adult-onset diseases such as ALS, and how diverse these motor neurons are in the adult stage of an animal.”
The study, “A molecular atlas of adult C. elegans motor neurons reveals ancient diversity delineated by conserved transcription factor codes,” was published February 29, 2024, in Cell Reports.
Surprisingly diverse motor neurons
Motor neurons are usually classified based on structural qualities like shape, where they live in the body, and what muscles they target. Kratsios and Jayson Smith, PhD, a postdoctoral scholar and first author of the study, worked with Seth Taylor and David Miller III from Vanderbilt University to use new single-cell RNA sequencing tools to analyze individual adult motor neurons in the roundworm. This kind of sequencing provides a snapshot of genetic expression in the cells, known as the transcriptome.
Using a sample of more than 13,000 cells, they classified the motor neurons into 29 distinct subclasses. The subclasses were organized based on distinct gene families. For example, expression of unique codes of either neuropeptides, proteins that stimulate neural activity outside the synapse, or transcription factors, proteins that control the rate and time of transcription of DNA into RNA, can distinguish one subclass from another. This molecular diversity of 29 subclasses is surprising, given that C. elegans has just 302 neurons total, including 75 motor neurons; this means that on average there are only two or three cells of any given motor neuron subclass in each worm. Future experiments are needed to discover the function of each subclass.
“Traditionally motor neurons are thought of as relatively simple. They just connect the central nervous system to the muscle, and then you get a muscle contraction,” Smith said. “Well, now we see there's all this molecular diversity by profiling the adult neurons.”
Deeply conserved organizing principles
Meanwhile, researchers at Stanford University performed single-cell RNA-sequencing on the mouse spinal cord and saw a similar level of diversity in its motor neurons. Smith and Kratsios compared the subclasses they discovered in the roundworm to the Stanford team’s findings from mice and saw that the same kind of organizing principles could apply, i.e., distinct codes of transcription factors can distinguish mouse motor neuron subclasses. This suggests that the way adult motor neurons are organized is conserved from ancient common ancestors, from simple invertebrate worms to complex modern mammals.
If we want to understand and treat adult-onset diseases of the nervous system, we must have a blueprint of how the system is built.
Paschalis Kratsios, PhD
Some of the transcription factors the team found that were important in adult cells are also important during early developmental stages, suggesting that the animals repurpose these genes in later stages of life. These developmental transcription factors are well-known, including homeodomain proteins that control large scale anatomical development, and HOX genes that dictate the head-tail arrangement of an animal. They also have similarities between mice and humans, meaning that the organizing principles seen in C. elegans can be extended further to humans.
First steps toward understanding disease progression
Smith said understanding how motor neuron subclasses are organized, specifically in adult cells, is crucial to understanding disease. In ALS, for example, sometimes the disease starts in the brainstem, affecting motor neurons controlling the patient’s speech and ability to swallow. It can also begin lower in the spinal cord and affect motor neurons critical for movement of the arms or legs.
“Adult-onset neurodegenerative diseases progress depending on the motor neuron subclass. So, understanding how some are resistant to disease and how others are susceptible to it is important,” Smith said. “Here we're taking the first steps toward understanding that question.”
The team has published their data online for use by other researchers at spinalcordatlas.org, along with mouse and human data so other researchers can compare and look for similarities between species. The RNA sequencing data is also combined with the C. elegans connectome, or a complete map of how each of the worm’s 302 neurons are connected to each other. Kratsios said he hopes to start studying the functionality and genetic mechanisms of them one-by-one next.
“The organizing principles and the molecular codes that we described for these 29 subclasses, maybe similar principles apply to the mouse brain or the human brain,” he said. "Imagine applying them to sensory circuits that are more complex, like the visual system or the auditory system. We can use it as a paradigm to understand and extrapolate basic science to address fundamental questions in the field of developmental neuroscience.”
The study, “A molecular atlas of adult C. elegans motor neurons reveals ancient diversity delineated by conserved transcription factor codes,” was supported by the National Institutes of Health (grants R21 NS108505, R01 Q12 NS118078, R01 NS116365, R01NS113559, R01NS100547, and R01NS106951). Additional authors include Jacob A. Blum and Aaron D. Gitler from Stanford University; Weidong Feng from the University of Chicago; and Rebecca Collings from Vanderbilt University.
This article was originally published on the BSD News website.